7. Utility Functions¶
-
MDSimsEval.utils.complement_residues(ligand_list_full, ligand_list_selected) Given a list of ligands and a list of ligands selected from the previous list, return the ligands included in the first but not the second list.
- Parameters
ligand_list_full – A list of
List[AnalysisActorClass]containing the full set of ligandsligand_list_selected – A list of
List[AnalysisActorClass]containing a subset of theligand_list_full
- Returns
A list of
List[AnalysisActorClass]containing the complement of the two sets above
-
MDSimsEval.utils.create_analysis_actor_dict(root_directory) Reads the simulations (topologies, trajectories and sasa.xvg, salts if available) and stores them in a dictionary structure. The dictionary structure called
analysis_actors_dictis the core structure that our functions take as an argument.- Example:
from MDSimsEval.utils import create_analysis_actor_dict analysis_actors_dict = create_analysis_actor_dict('path_to_data_directory/') analysis_actors_dict['Agonists'][0].info() Out: <<< Info of 5-MeOT >>> Number of Frames: 2500 Number of Atoms: 4743 Number of Residues: 291
- Parameters
root_directory (str) – The path of the input directory having the expected structure on the documentation
- Returns
analysis_actors_dict:
Dict( "Agonists": List[AnalysisActor.class] "Antagonists": List[AnalysisActor.class] )
-
MDSimsEval.utils.render_corr_df(corr_df, filepath, reversed=False) Renders and saves a correlation heatmap which is visually interpretable.
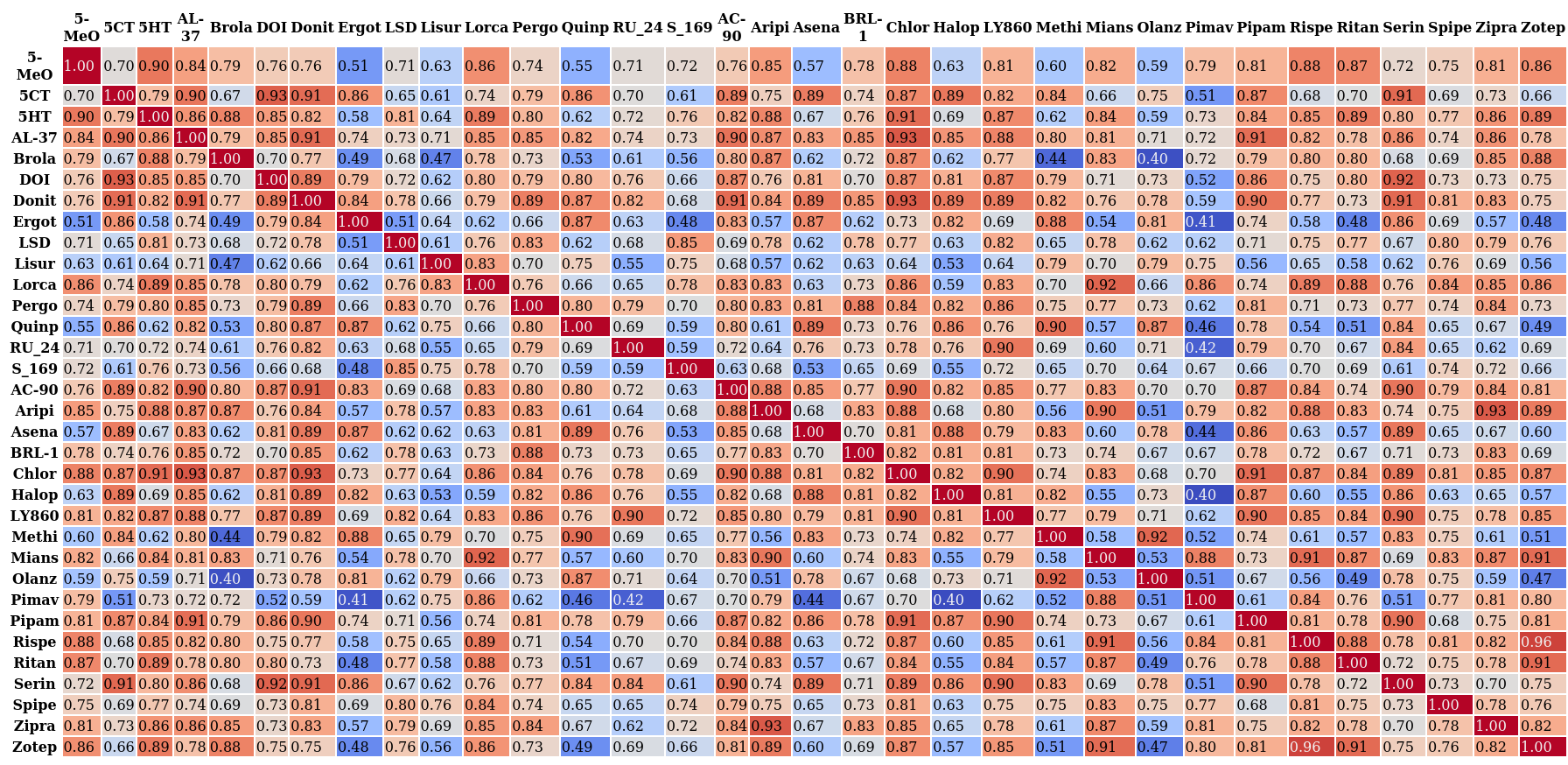
Correlation heatmap, click for higher resolution.¶
Note
In order to save the image as
.pngyou must installwkhtmltopdfviasudo apt-get install wkhtmltopdfon your machine. Else the output will be in a.htmlfile and can be viewed using any browser.- Parameters
corr_df (pd.DataFrame) – A DataFrame of pairwise correlations
filepath (str) – The full path the heatmap save location eg.
/dir1/dir2/name_of_filereversed (boolean) – If you want the coolwarm colormap to be inversed