2. Reading the simulations¶
This doc page will guide you through preparing your data in the package expected structure and present a function for reading and storing them.
2.1. Directory Structure¶
Our reader function expects a specific directory structure.
input_directory/
Agonists/
Ligand1_/
trajectory_.xtc
topology_.pdb
sasa.xvg *
salts/ *
Ligand2_
.
.
.
Antagonists/
.
.
.
On the above structure everything followed by an underscore _ can have a different name.
Agonists and Antagonists were the class labels of my use case. Currently, you must follow the same names as
class labels. Should fixed, so as class label is also a variable.
Note
sasa.xvg and salts/ are explained in SASA and Salt Bridges.
As an example this was my actual input directory named New_AI_MD:
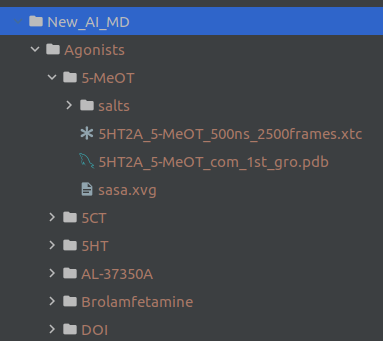
2.2. Reading Function¶
Reads the simulations (topologies, trajectories and sasa.xvg, salts if available) and stores them in a dictionary
structure. The dictionary structure called analysis_actors_dict is the core structure that our functions
take as an argument.
- Example:
- param root_directory
The path of the input directory having the expected structure on the documentation
- type root_directory
str
- returns
analysis_actors_dict:
Dict( "Agonists": List[AnalysisActor.class] "Antagonists": List[AnalysisActor.class] )
2.3. SASA and Salt Bridges¶
These two features have a different way of being calculated outside of this package.
Concerning SASA we must first create a sasa.xvg file for each simulation and keep it next to the topology and the
trajectory. The GROMACS command is:
gmx sasa -f trajecotry.xtc -s topology.pdb -o sasa.xvg
# Select Group 1 (Protein)
Concerning the salt bridges you must use a VMD plugin and
copy the whole outputted directory named salts/ next to the trajectory files.
Note
Currently no analysis functions uses salt bridges in this package.